



Research theme
The molecular basis of early events in plant development such as patterning the apical meristem and specifying organ identity have been studied in detail by several laboratories worldwide. Yet how this regional information is translated into the mature organ of specific shape and size is poorly understood. For example, it is not clear how various cell types differentiate in a coordinated manner to shape up the final structure of leaf, petal, or fruit. Our laboratory is interested in studying the genetic basis of organ growth and development in plants.
Research Profile
How shape evolves during organ growth is an important question in developmental biology. We address this by analysing growth of plant organs such as leaves in the planar dimension. Leaves are initiated as mound of cells called primordium at the flank of the shoot apical meristem, a stem cell niche that generates all above-ground parts of a plant. At the inception, growth is uniform throughout the primordium. In certain species such as Arabidopsis, as development progresses, growth stops towards the distal end while the base continues to grow, thus creating a base-to-tip polar growth pattern. Analysis of leaf growth in a large number of diverse plant species using the law of allometry revealed that, although leaves of some species grow from the base, others grow from the tip, while yet another group of leaves grows with no apparent polarity, thereby generating a natural diversity in growth polarity. The plant-specific microRNA miR396 regulates this growth diversity.
Despite their anisometric growth as described above, how leaves retain their surface flatness is still an open question. Studies from several laboratories including ours have identified two groups of genes that control surface curvature: one group codes for miR319-regulated TEOSINTE BRANCHED1, CYCLOIDEA, PROLIFERATING CELL FACTORS (also called JAW-TCP) transcription factors that suppress marginal growth, while the other group encodes proteins involved in proteasome-mediated protein degradation and suppress growth at the centre. Mutations in either group result in the loss of leaf flatness. Genetic interaction studies reveal that these two genetic pathways function independently.
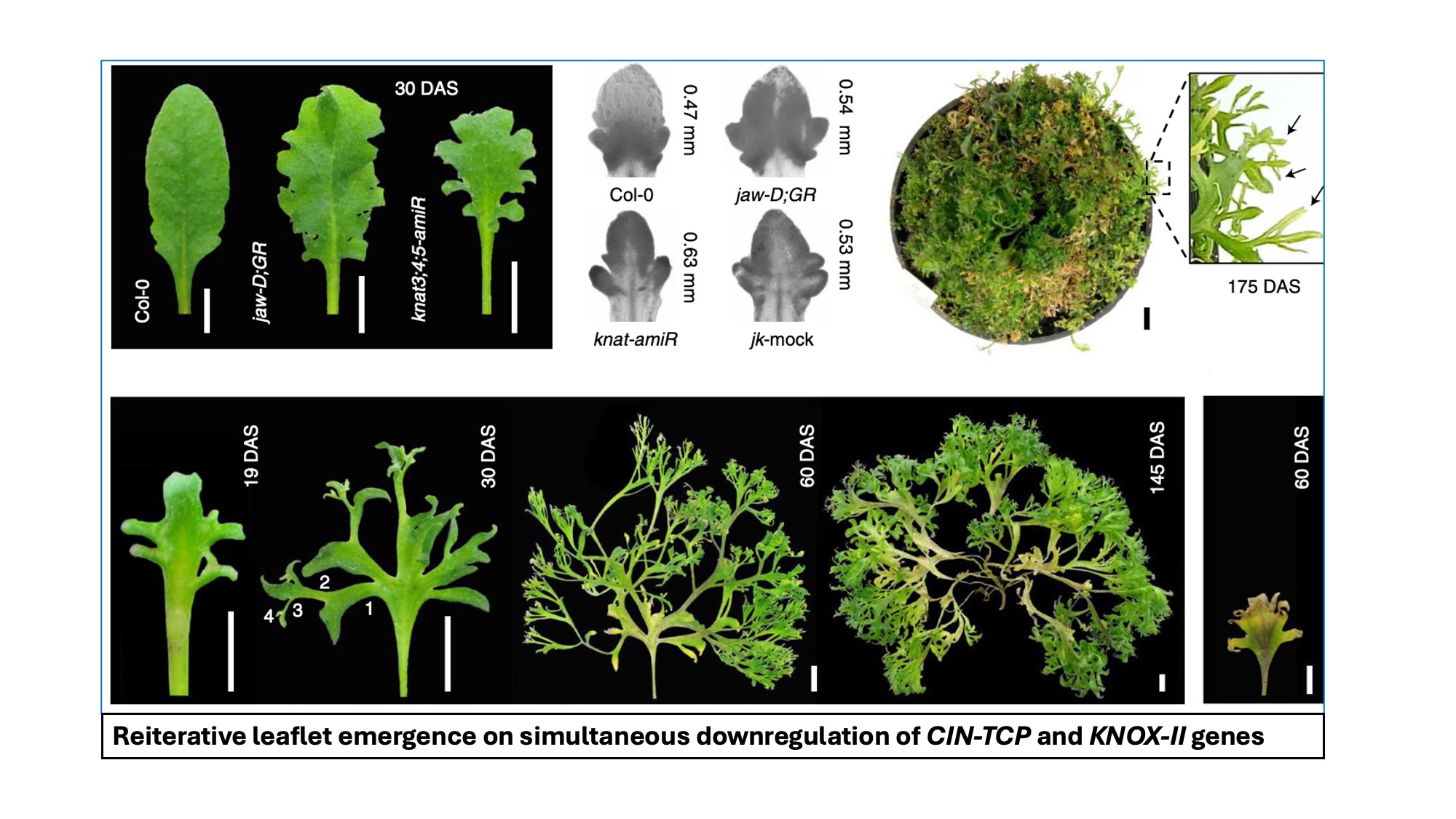
Leaves show extensive shape diversity and are broadly divided into two forms; simple leaves with intact lamina and compound leaves with lamina dissected into leaflets. The mechanistic basis of margin dissection and leaflet initiation has been inferred primarily by analysing compound-leaf architecture, and thus whether the intact lamina of simple leaves has the potential to initiate leaflets upon endogenous gene inactivation remains unclear. We have shown that the JAW-TCP transcription factors activate the class II KNOTTED1-LIKE (KNOX-II) genes, and the JAW-TCP and KNOX-II proteins together redundantly suppress leaflet initiation in simple leaves. Simultaneous downregulation of JAW-TCP and KNOX-II in Arabidopsis leads to the reactivation of the stemness genes KNOX-I and CUPSHAPED COTYLEDON (CUC) and triggers ectopic organogenesis, eventually converting the simple lamina to a super-compound form that appears to initiate leaflets indefinitely. Thus, a conserved developmental mechanism promotes simple leaf architecture in which JAW-TCP–KNOX-II forms a strong differentiation module that suppresses the KNOX-I-CUC network and leaflet initiation.
These findings help us understand how genes regulate biological shape. We are currently studying regulation of leaf shape in other species to test whether the molecular mechanisms underlying organ geometry in evolutionarily conserved.
Faculty Profile
PhD: National Centre for Biological Science, TIFR Centre, Bangalore, India
Postdoc: John Innes Centre, Norwich, UK

Email : abhishekgupt@iisc.ac.in
Designation : Phd Student
Category : Plant and Developmental Biology
Email : aliaktaras@iisc.ac.in
Designation : Phd Student
Category : Plant and Developmental Biology

Email : anjanahegde@iisc.ac.in
Designation : Phd Student
Category : Plant and Developmental Biology

Email : anuragsharma@iisc.ac.in
Designation : Phd Student
Category : Plant and Developmental Biology

Email : arpitac@iisc.ac.in
Designation : Phd Student
Category : Plant and Developmental Biology

Email : binayakumar@iisc.ac.in
Designation : M.Sc Dissertation Student
Category : Plant and Developmental Biology

Email : bonanidas@iisc.ac.in
Designation : Phd Student
Category : Plant and Developmental Biology

Email : nargislaha@iisc.ac.in
Designation : postdoc
Category : Plant and Developmental Biology

Email : priyankamish@iisc.ac.in
Designation : postdoc
Category : Plant and Developmental Biology

Email : yadukrishnan@iisc.ac.in
Designation : postdoc
Category : Plant and Developmental Biology

Email : gulfisham@iisc.ac.in
Designation : Phd Student
Category : Plant and Developmental Biology
Email : bsharath@iisc.ac.in
Designation : Phd Student
Category : Plant and Developmental Biology
Email : somsreeroy@iisc.ac.in
Designation : Phd Student
Category : Plant and Developmental Biology

Email : vishwadeepm@iisc.ac.in
Designation : Phd Student
Category : Plant and Developmental Biology
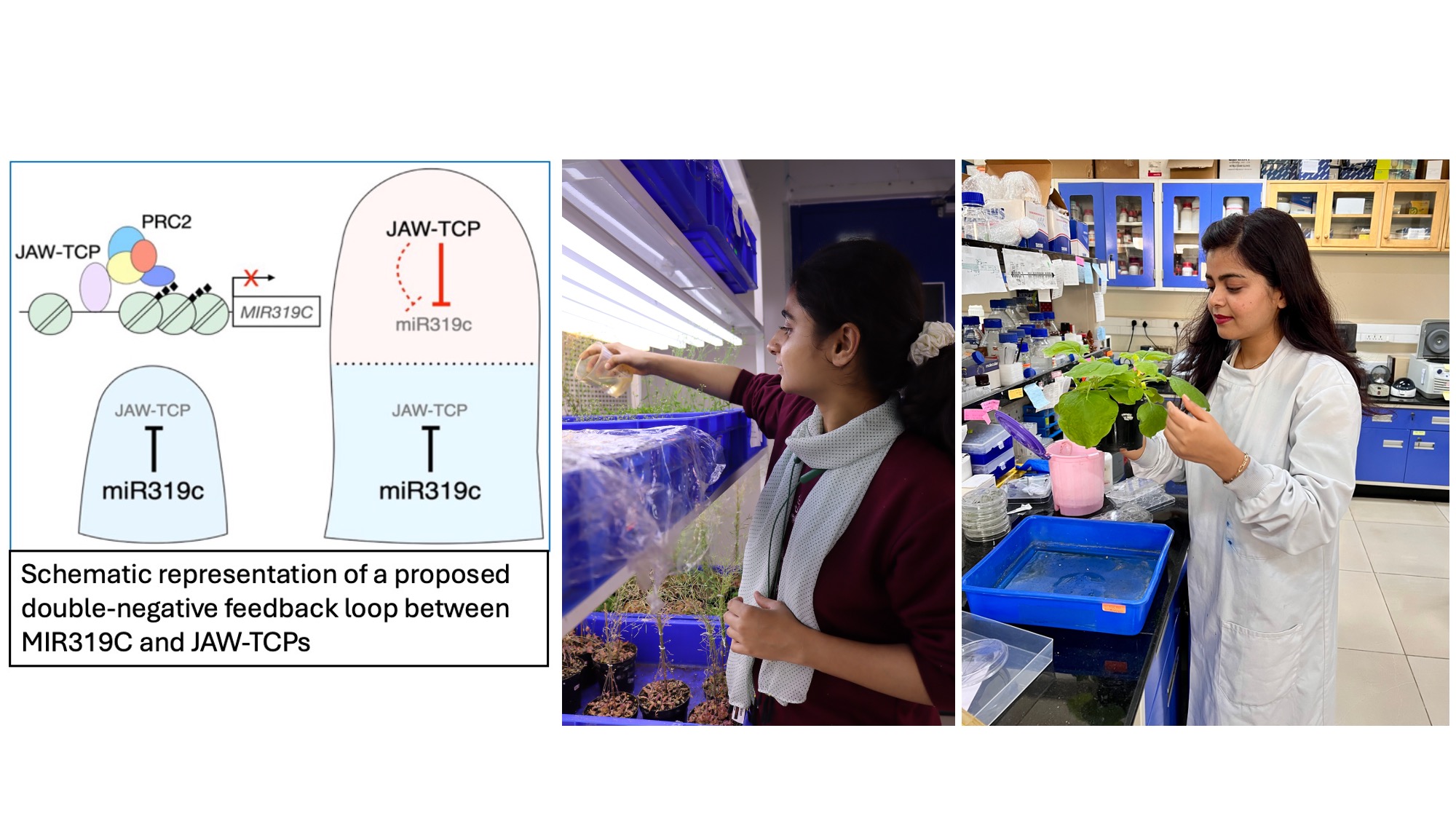
The microRNA miR319 and its target JAW-TCP transcription factors regulate the proliferation-to-differentiation transition of leaf pavement cells in diverse plant species. In young Arabidopsis leaf primordia, JAW-TCPs are detected towards the distal region whereas the major mRNA319-encoding gene MIR319C, is expressed at the base. Little is known about how this complementary expression pattern of MIR319C and JAW-TCPs is generated. Here, we show that MIR319C is initially expressed uniformly throughout the incipient primordia and is later abruptly down-regulated at the distal region, with concomitant distal appearance of JAW-TCPs, when leaves grow to ~100 μm long. Loss of JAW-TCPs causes distal extension of the MIR319C expression domain, whereas ectopic TCP activity restricts MIR319C more proximally. JAW-TCPs are recruited to and are capable of depositing histone H3K27me3 repressive marks on the MIR319C chromatin. JAW-TCPs fail to repress MIR319C in transgenic seedlings where the TCP-binding cis-elements on MIR319C are mutated, causing miR319 gain-of-function-like phenotype in the embryonic leaves. Based on these results, we propose a model for growth patterning in leaf primordia wherein MIR319C and JAW-TCPs repress each other and divide the uniformly growing primordia into distal differentiation zone and proximal proliferation domain.
Angiosperm leaves show extensive shape diversity and are broadly divided into two forms; simple leaves with intact lamina and compound leaves with lamina dissected into leaflets. The mechanistic basis of margin dissection and leaflet initiation has been inferred primarily by analysing compound-leaf architecture, and thus whether the intact lamina of simple leaves has the potential to initiate leaflets upon endogenous gene inactivation remains unclear. Here, we show that the CINCINNATA-like TEOSINTE BRANCHED1, CYCLOIDEA, PROLIFERATING CELL FACTORS (CIN-TCP) transcription factors activate the class II KNOTTED1-LIKE (KNOX-II) genes and the CIN-TCP and KNOX-II proteins together redundantly suppress leaflet initiation in simple leaves. Simultaneous downregulation of CIN-TCP and KNOX-II in Arabidopsis leads to the reactivation of the stemness genes KNOX-I and CUPSHAPED COTYLEDON (CUC) and triggers ectopic organogenesis, eventually converting the simple lamina to a super-compound form that appears to initiate leaflets indefinitely. Thus, a conserved developmental mechanism promotes simple leaf architecture in which CIN-TCP–KNOX-II forms a strong differentiation module that suppresses the KNOX-I-CUC network and leaflet initiation.
The ubiquitin-mediated proteasomal pathway regulates diverse cellular processes in plants by rapidly degrading target proteins, including the repressors of hormone signaling. Though ubiquitin proteases play a key role in this process by cleaving polyubiquitin chains to monomers, their function has not been studied in detail by mutational analysis. Here, we show that mutation in TARANI/UBIQUITIN-SPECIFIC PROTEASE14 (TNI/UBP14) leads to reduced auxin response and widespread auxin-related phenotypic defects in Arabidopsis (Arabidopsis thaliana). In a tni partial loss-of-function mutant that was originally isolated based on altered leaf shape, activity of the auxin-responsive reporters DR5::GUS, DR5::nYFP, and IAA2::GUS was reduced. Genetic interaction studies suggest that TNI is involved in auxin signaling and acts alongside TIR1, ARF7, and AUX1. Map-based cloning identified TNI as UBP14. Inefficient splicing of the mutant TNI transcript resulted in the formation of an inactive UBP14 protein, which led to accumulation of polyubiquitin chains and excess polyubiquitinated proteins in the mutant. In addition to the reduced auxin response, increased levels of DII:VENUS, IAA18:GUS, and HS::AXR3-NT:GUS were also observed in tni, perhaps due to inefficient polyubiquitin hydrolysis and proteasome-mediated degradation. Together, our study identifies a function for TNI/UBP14 in the auxin response through ubiquitin recycling.
Cells in organ primordia undergo active proliferation at an early stage to generate sufficient number, before exiting proliferation and entering differentiation. However, how the actively proliferating cells are developmentally reprogrammed to acquire differentiation potential during organ maturation is unclear. Here, we induced a microRNA-resistant form of TCP4 at various developmental stages of Arabidopsis leaf primordium that lacked the activity of TCP4 and its homologues and followed its effect on growth kinematics. By combining this with spatio-temporal gene expression analysis, we show that TCP4 commits leaf cells within the transition zone to exit proliferation and enter differentiation. A 24-hour pulse of TCP4 activity was sufficient to impart irreversible differentiation competence to the actively dividing cells. A combination of biochemical and genetic analyses revealed that TCP4 imparts differentiation competence by promoting auxin response as well as by directly activating HAT2, a HD-ZIP II transcription factor-encoding gene that also acts downstream to auxin response. Our study offers a molecular link between the two major organ maturation factors, CIN-like TCPs and HD-ZIP II transcription factors and explains how TCP activity restricts the cell number and final size in a leaf.
Trichomes are the first line of defense on the outer surface of plants against biotic and abiotic stresses. Because trichomes on leaf surfaces originate from the common epidermal progenitor cells that also give rise to pavement cells and stomata, their density and distribution are under strict genetic control. Regulators of trichome initiation have been identified and incorporated into a biochemical pathway wherein an initiator complex promotes trichome fate in an epidermal progenitor cell, while an inhibitor complex suppresses it in the neighboring cells. However, it is unclear how these regulator proteins, especially the negative regulators, are induced by upstream transcription factors and integrated with leaf morphogenesis. Here, we show that the Arabidopsis (Arabidopsis thaliana) class II TCP proteins activate TRICHOMELESS1 (TCL1) and TCL2, the two established negative regulators of trichome initiation, and reduce trichome density on leaves. Loss-of-function of these TCP proteins increased trichome density whereas TCP4 gain-of-function reduced trichome number. TCP4 binds to the upstream regulatory elements of both TCL1 and TCL 2 and directly promotes their transcription. Further, the TCP-induced trichome suppression is independent of the SQUAMOSA PROMOTER BINDING PROTEIN LIKE family of transcription factors, proteins that also reduce trichome density at later stages of plant development. Our work demonstrates that the class II TCP proteins couple leaf morphogenesis with epidermal cell fate determination.
Organ elaboration in plants occurs almost exclusively by an
increase in cell number and size. Leaves, the planar lateral
appendages of plants, are no exception. Forward and reverse
genetic approaches have identified several genes whose role in
leaf morphogenesis has been inferred from their primary effect
on cell number and size, thereby distinguishing them as either
promoters or inhibitors of cell proliferation and expansion.
While such classification is useful in studying size control, a
similar link between genes and shape generation is poorly
understood. Computational modelling can provide a
conceptual framework to re-evaluate the known genetic
information and assign specific morphogenetic roles to the
transcription factor-encoding genes. Here we discuss recent
advances in our understanding of the roles of transcription
factors in the planar growth of leaf lamina in two orthogonal
dimensions.
Leaves are the most conspicuous planar organs in plants, designed for efficient capture of sunlight and its conversion to energy that is channeled into sustaining the entire biosphere. How a few founder cells derived from the shoot apical meristem give rise to diverse leaf forms has interested naturalists and developmental biologists alike. At the heart of leaf morphogenesis lie two simple cellular processes, division and expansion, that are spatially and temporally segregated in a developing leaf. In leaves of dicot model species, cell division occurs predominantly at the base, concomitant with the expansion and differentiation of cells at the tip of the lamina that drives increase in leaf surface area. The timing of the transition from one cell fate (division) to the other (expansion) within a growing leaf lamina is a critical determinant of final leaf shape, size, complexity and flatness. The TCP proteins, unique to plant kingdom, are sequence-specific DNA-binding transcription factors that control several developmental and physiological traits. A sub-group of class II TCPs, called CINCINNATA-like TCPs (CIN-TCPs henceforth), are key regulators of the timing of the transition from division to expansion in dicot leaves. The current review highlights recent advances in our understanding of how the pattern of CIN-TCP activity is translated to the dynamic spatio-temporal control of cell-fate transition through the transactivation of cell-cycle regulators, growth-repressing microRNAs, and interactions with the chromatin remodeling machinery to modulate phytohormone responses. Unravelling how environmental inputs influence CIN-TCP-mediated growth control is a challenge for future studies.
In plants, endogenous and environmental signals such as light control the timing of the transition to flowering. Two phytochrome B-interacting transcription factors, VASCULAR PLANT ONE-ZINC FINGER1 (VOZ1) and VOZ2, redundantly promote flowering in Arabidopsis (Arabidopsis thaliana). In the voz1 voz2 mutant, the expression of FLOWERING LOCUS C (FLC) was up-regulated and that of FLOWERING LOCUS T (FT) was down-regulated, which was proposed to be the cause of late flowering in voz1 voz2. However, the detailed mechanism by which the VOZ genes promote flowering is not well understood. Here, we show that neither the reduced FT expression nor the late-flowering phenotype of voz1 voz2 is suppressed in the voz1 voz2 flc triple mutant. Genetic interaction experiments between voz1 voz2 and constans-2 (co-2) mutants reveal that the VOZs and CO work in the same genetic pathway. Using in vitro pull-down, electrophoretic mobility shift, and bimolecular fluorescence complementation assays, we show that VOZ1 and VOZ2 interact with CO. The voz1 voz2 35S::CO:YFP plants show suppression of the early-flowering phenotype induced by CO overexpression, suggesting that CO requires VOZ for the induction of flowering. Determination of the VOZ consensus-binding site followed by genome-wide sequence analysis failed to identify any VOZ-binding sites near known flowering time genes. Together, these results indicate that the VOZ genes regulate flowering primarily through the photoperiod pathway, independent of FLC, and suggest that VOZs modulate CO function to promote flowering.
Trichomes are the first cell type to be differentiated during the morphogenesis of leaf epidermis and serve as an ideal model to study cellular differentiation. Many genes involved in the patterning and differentiation of trichome cells have been studied over the past decades, and the majority of these genes encode transcription factors that specifically regulate epidermal cell development. However, the upstream regulators of these genes that link early leaf morphogenesis with cell type differentiation are less studied. The TCP proteins are the plant-specific transcription factors involved in regulating diverse aspects of plant development including lateral organ morphogenesis by modulating cell proliferation and differentiation. Here, we show that the miR319-regulated class II TCP proteins, notably TCP4, suppress trichome branching in Arabidopsis leaves and inflorescence stem by direct transcriptional activation of GLABROUS INFLORESCENCE STEMS (GIS), a known negative regulator of trichome branching. The trichome branch number is increased in plants with reduced TCP activity and decreased in the gain-of-function lines of TCP4. Biochemical analyses show that TCP4 binds to the upstream regulatory region of GIS and activates its expression. Detailed genetic analyses show that GIS and TCP4 work in same pathway and GIS function is required for TCP4-mediated regulation of trichome differentiation. Taken together, these results identify a role for the class II TCP genes in trichome differentiation, thus providing a connection between organ morphogenesis and cellular differentiation.
Cell expansion is an essential process in plant morphogenesis and is regulated by the coordinated action of environmental stimuli and endogenous factors, such as the phytohormones auxin and brassinosteroid. Although the biosynthetic pathways that generate these hormones and their downstream signaling mechanisms have been extensively studied, the upstream transcriptional network that modulates their levels and connects their action to cell morphogenesis is less clear. Here, we show that the miR319-regulated TCP (TEOSINTE BRANCHED1, CYCLODEA, PROLIFERATING CELL FACTORS) transcription factors, notably TCP4, directly activate YUCCA5 transcription and integrate the auxin response to a brassinosteroid-dependent molecular circuit that promotes cell elongation in Arabidopsis thaliana hypocotyls. Furthermore, TCP4 modulates the common transcriptional network downstream to auxin-brassinosteroid signaling, which is also triggered by environmental cues, such as light, to promote cell expansion. Our study links TCP function with the hormone response during cell morphogenesis and shows that developmental and environmental signals converge on a common transcriptional network to promote cell elongation.
Lateral appendages often show allometric growth with a specific growth polarity along the proximo-distal axis. Studies on leaf growth in model plants have identified a basipetal growth direction with the highest growth rate at the proximal end and progressively lower rates toward the distal end. Although the molecular mechanisms governing such a growth pattern have been studied recently, variation in leaf growth polarity and, therefore, its evolutionary origin remain unknown. By surveying 75 eudicot species, here we report that leaf growth polarity is divergent. Leaf growth in the proximo-distal axis is polar, with more growth arising from either the proximal or the distal end; dispersed with no apparent polarity; or bidirectional, with more growth contributed by the central region and less growth at either end. We further demonstrate that the expression gradient of the miR396-GROWTH-REGULATING FACTOR module strongly correlates with the polarity of leaf growth. Altering the endogenous pattern of miR396 expression in transgenic Arabidopsis thaliana leaves only partially modified the spatial pattern of cell expansion, suggesting that the diverse growth polarities might have evolved via concerted changes in multiple gene regulatory networks.
Plant organs are initiated as primordial outgrowths, and require controlled cell division and differentiation to achieve their final size and shape. Superimposed on this is another developmental program that orchestrates the switch from vegetative to reproductive to senescence stages in the life cycle. These require sequential function of heterochronic regulators. Little is known regarding the coordination between organ and organismal growth in plants. The TCP gene family encodes transcription factors that control diverse developmental traits, and a subgroup of class II TCP genes regulate leaf morphogenesis. Absence of these genes results in large, crinkly leaves due to excess division, mainly at margins. It has been suggested that these class II TCPs modulate the spatio-temporal control of differentiation in a growing leaf, rather than regulating cell proliferation per se. However, the link between class II TCP action and cell growth has not been established. As loss-of-function mutants of individual TCP genes in Arabidopsis are not very informative due to gene redundancy, we generated a transgenic line that expressed a hyper-activated form of TCP4 in its endogenous expression domain. This resulted in premature onset of maturation and decreased cell proliferation, leading to much smaller leaves, with cup-shaped lamina in extreme cases. Further, the transgenic line initiated leaves faster than wild-type and underwent precocious reproductive maturation due to a shortened adult vegetative phase. Early senescence and severe fertility defects were also observed. Thus, hyper-activation of TCP4 revealed its role in determining the timing of crucial developmental events, both at the organ and organism level.
The TCP transcription factors control multiple developmental traits in diverse plant species. Members of this family share an ∼60-residue-long TCP domain that binds to DNA. The TCP domain is predicted to form a basic helix-loop-helix (bHLH) structure but shares little sequence similarity with canonical bHLH domain. This classifies the TCP domain as a novel class of DNA binding domain specific to the plant kingdom. Little is known about how the TCP domain interacts with its target DNA. We report biochemical characterization and DNA binding properties of a TCP member in Arabidopsis thaliana, TCP4. We have shown that the 58-residue domain of TCP4 is essential and sufficient for binding to DNA and possesses DNA binding parameters comparable to canonical bHLH proteins. Using a yeast-based random mutagenesis screen and site-directed mutants, we identified the residues important for DNA binding and dimer formation. Mutants defective in binding and dimerization failed to rescue the phenotype of an Arabidopsis line lacking the endogenous TCP4 activity. By combining structure prediction, functional characterization of the mutants, and molecular modeling, we suggest a possible DNA binding mechanism for this class of transcription factors.
Considerable progress has been made in identifying the targets of plant microRNAs, many of which regulate the stability or translation of mRNAs that encode transcription factors involved in development. In most cases, it is unknown, however, which immediate transcriptional targets mediate downstream effects of the microRNA-regulated transcription factors. We identified a new process controlled by the miR319-regulated clade of TCP (TEOSINTE BRANCHED/CYCLOIDEA/PCF) transcription factor genes. In contrast to other miRNA targets, several of which modulate hormone responses, TCPs control biosynthesis of the hormone jasmonic acid. Furthermore, we demonstrate a previously unrecognized effect of TCPs on leaf senescence, a process in which jasmonic acid has been proposed to be a critical regulator. We propose that miR319-controlled TCP transcription factors coordinate two sequential processes in leaf development: leaf growth, which they negatively regulate, and leaf senescence, which they positively regulate.

nishagupta@iisc.ac.in

amrita.bhu08@gmail.com

shyam.gundu@kaust.edu.sa

monalishar@iisc.ac.in

preethis@iisc.ac.in

vbattala@salk.edu

naveenalanga@iisc.ac.in