Research
Development of multicellular eukaryotes can be controlled by master regulators with cascading effects on cell and tissue fate. In higher plants, the embryonically established Shoot Apical Meristem (SAM) is critical growing center that maintains a pool of stem cell and is the source of new cells for all aerial tissues e.g., leaves, branches with new axillary meristems that can form secondary growth axis. In flowering plants, transition to reproductive phase of growth is reflected by a dramatic change in the fate of organs that emerge from the apical meristem that now produce flowers with specialized organs. While flowers—the progenitors of fruits, seeds, and grains—have stereotypic concentric pattern of specific floral organs there is again an astonishing diversity in the time taken for transition to flowering, in size, shape, position, and tissue differentiation patterns in floral organs.
Genetic regulators of flowering in rice: Rice, is a genetically tractable cereal-grass model, that produces modified flowers (called florets) on higher-order inflorescence branches (Figure 1A). Additionally, florets in rice and other cereals, have unique outer floral organs called lemma, palea that fully enclose the flower. Rice floret opening is temporally controlled and allows for anthesis, and floret closure ensures proper seed setting. These functions are conferred by specialized organs called lodicules that are asymmetrically positioned and are unusually short and fleshy organs that act as mechanical levers (Figure 1A). The homologies of these organs to dicot sepal and petal of dicot flowers organs are debated. We are studying how evolutionarily conserved regulators of angiosperm flower specification and organ differentiation confer different architectures of the rice inflorescence and of floret organs. Understanding rice genes that regulate flowering and fertility is important for knowledge on developmental gene regulation and for crop breeding. We use and innovate functional genetics (RNAi, genome-edited loss of function mutants, transgenics) and multi-omics to decipher functions for transcription factors that control developmental gene networks for flowering and fertility.
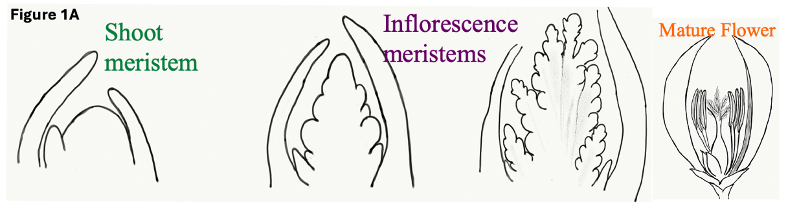
RFL, rice LFY, and control of meristem fate:
In Arabidopsis LFY, a bHTH motif containing transcription factor, triggers the floral program on the flanks of the apical inflorescence meristem; yet as LFY orthologs in other species have species-specific expression patterns this opens several conceptual questions on its role in diverse species. Our work on rice RFL showed unique cis- enhancers drive its dynamic expression in inflorescence branch meristems, which we have shown is important for branching. We have studied the consequences of RFL knockdown by RNAi and in a genome edited mutant with a partial loss of function mutant that points to novel species-specific roles in several rice meristems. Loss of RFL causes show delayed transition to flowering, poor inflorescence branching and very interesting recent data point to likely roles adventitious root formation.
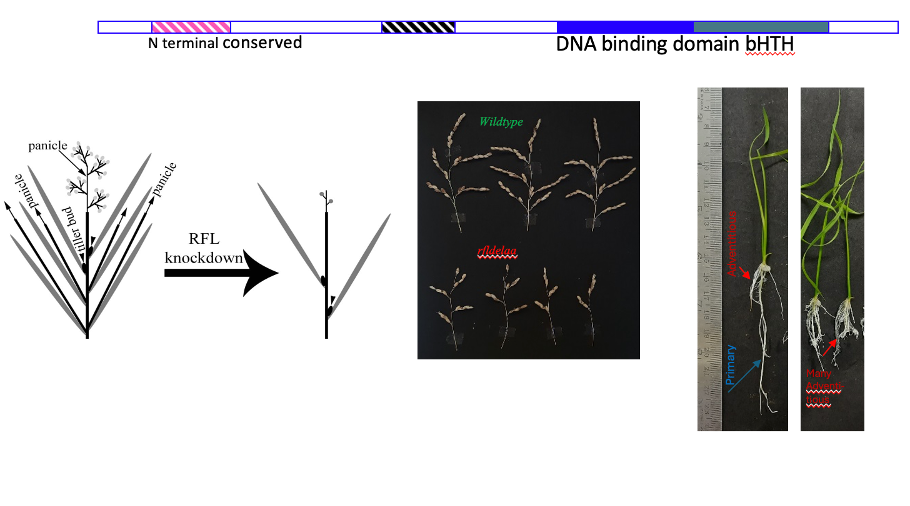
We are teasing out the molecular basis of these varied developmental effects of RFL by employing genetic (mutants, transgenics with induced overexpression/ knockdown), genomic (ChIP-seq, RNA-seq), biochemical (DNA-protein binding assays) and imaging tools (for cell division, mapping auxin rich cells) to decipher direct and indirectly regulated RFL gene targets. Broad questions being investigated are:
- What is the transcriptome of different meristems in rice? Are there developmental pre-patterns?
- How do RFL and its target genes control meristem cell proliferation vs. differentiation
The outcomes of these studies will identify developmental and cellular pathways common to all meristems, as compared to those that are meristems-specific differentiation programs.
OsMADS1 a regulatory hub for rice floral meristem formation and differentiation
The stereotypic position of floral organs on the developing floral meristem is largely determined by the combinatorial complexes of MADS-domain transcription factors. Rice florets are made on final short branch called the spikelet. To decipher snapshots of developmentally co-regulated genes during rice floret meristem establishment, organ primordia patterning and organ differentiation we determined transcriptome profiles from wild-type inflorescence tissues binned to represent these three broad development pools. In these three developmental pools, co-expressed genes were identified that will guide our ongoing work on meristem stem cell maintenance vs. stem cell termination and organ specification. Further, our current research explores how OsMADS1, a MADS factor belonging to the SEP clade, controls formation of determinate floret meristem with fixed number of organs with distinctive organs. Specifically, we generated a null osmads1 knockout mutant to identify that loss of this single gene function abrogates entire floret formation program. In the mutant floret meristem is abnormal with an excess of organs all of which are malformed with loss of organ identity. Using this genetic resource, some broad questions we are addressing are:
- How does OsMADS1 control the spatial and temporal activity of stem cells in incipient and developing floret meristems?
- What is the temporal cascade of OsMADS1 gene regulatory networks in developing florets? Do specific downstream targets control differentiation pathways?
- Does OsMADS1 regulated floret establishment and maintenance involve epigenetic control of development?
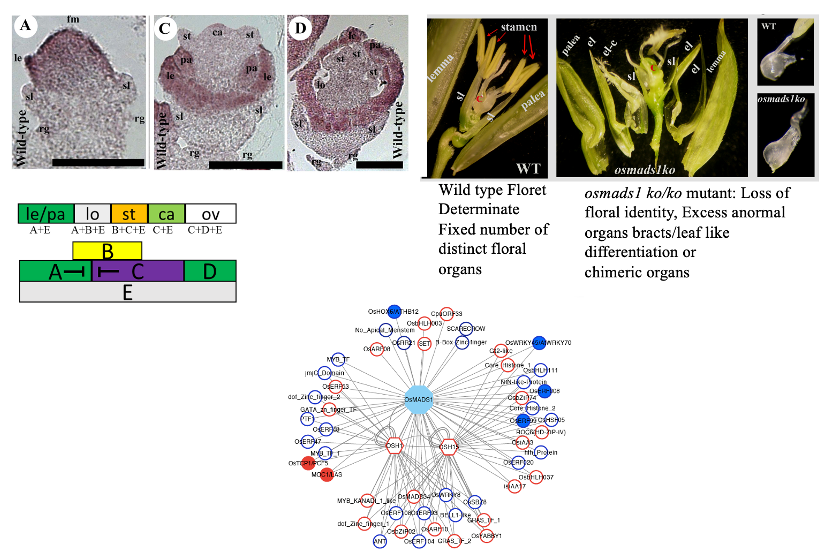
The OsMADS1 transcriptome and its chromatin occupancy are giving leads on its temporally varied effects on downstream transcription factors (homeodomain, Basic HLH domain), chromatin modifiers (Jumonji domain, Trx group factor), cell proliferation, and hormone signaling. These snapshots of OsMADS1 driven gene networks give leads on pathways that ensure floret development and successful reproduction. We are actively pursuing some key bHLH transcription factor downstream target genes (OsSPT, OsHEC), ETT class of auxin response factors and Trithorax group factor to delineate their contributions in floret meristems and organ primordia.
Unravelling unequal redundancies and novel roles for rice PISTILLATA (PI) clade organ patterning genes OsMADS2 and OsMADS4
Lodicules are a pair of unique second whorl small fleshy organs in rice and cereal florets that are regarded as the eudicot petal analogue. Their small size, cup shape, and asymmetric position on one half whorl are strikingly different from flat, full whorl of large exposed petals of eudicots. The unique morphological and functional properties of lodicules direct their role in the timed opening and subsequent closure of the rice floret, thus permitting stamen emergence and cross-pollination. Here we are investigating the developmental functions of the rice floral organ patterning PISTILLATA (PI)paralogs; OsMADS2 and OsMADS4 whose orthologs in Arabidopsis and eudicot models are required for petal and stamen development. How OsMADS2 influences organ size, shape and tissue differentiation in lodicules is being explored using mutants, transgenic complementation assays, and functional genomics. Additionally, our ongoing work with genetic null osmads2 mutant and lines with doubly perturbed mutant (osmads2 dsRNAiOsMADS4) are pointing to roles in many unanticipated aspects of flowering (flowering time, floral meristem size) and in the suppression of parthenocarpy.
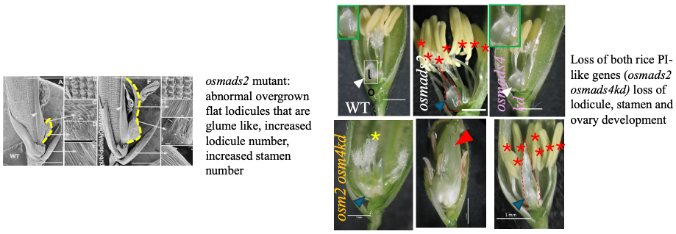
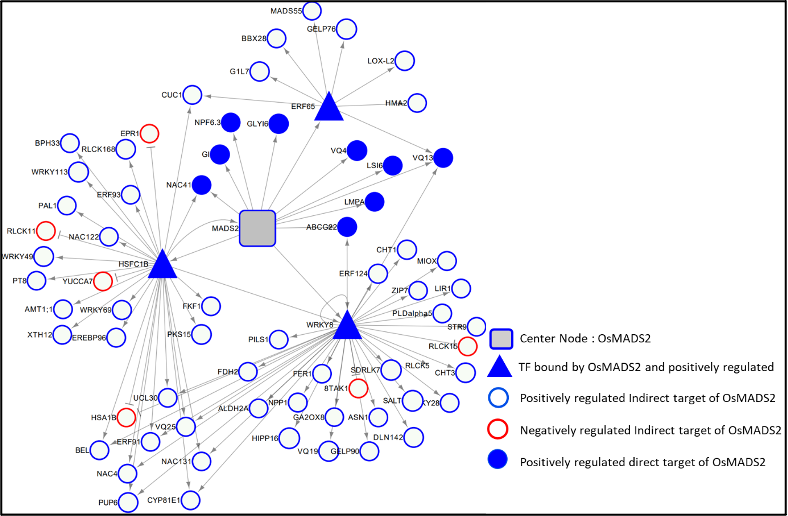
Using these genetic resources, some broad questions we are addressing are:
- How does OsMADS2 control the spatial and temporal pattern of cell division vs. elongation in organ primordia to control organ size shape and tissue differentiation?
- What are its specific downstream targets that control the unique dispersed vascular pattern in lodicules?
- How do OsMADS2 and OsMADS4 help co-ordinate ovary wall growth with embryo development?
The insights on developmental pathways and target genes regulated by OsMADS2 that we have recently deciphered from RNA-Seq and ChIP-Seq analyses are being extended to provide deeper molecular insights conserved vs. unique roles for these rice PI-clade transcription factors.
Pre-mRNA splicing and links to regulated gene expression and cellular processes: RNA splicing, an essential step in eukaryotic gene expression, generates functional RNAs from a gene's primary transcript. Splicing requires precise excision of non-functional intronic segments, and this can expand the complexity of expressed information from any genome. Defects in splicing cause diseases and developmental disorders in plant and animal systems. Our past research on fission yeast factors, Prp18, Slu7 and Prp16, deciphered their intron-context-dependent roles for accurate splice-site selection and for alternative splice isoforms under diverse growth and stress conditions. More recently, using the short intron-rich pathogenic budding yeast model Cryptococcus neoformans we have deciphered how splicing factors fine-tune splicing and gene expression to control cell cycle progression, nuclear position during mitosis and faithful chromosome segregation. As the short intron and multiple introns/ transcript gene architecture in this genome resembles those some other pathogens, in higher plants, nematodes, and certain higher eukaryotes our findings preview how post-transcriptional mechanisms may fine-tune developmental gene expression.
Group Member

Amartya Sarkar
Email : amartyas@iisc.ac.in
Designation : Phd Student
Category : Plant and Developmental Biology

Heeba Anjum
Email : heebaanjum@iisc.ac.in
Designation : Phd Student
Category : Plant and Developmental Biology

Kirti Ravat
Email : kirtiravat@iisc.ac.in
Designation : Phd Student
Category : Plant and Developmental Biology

MANENDRA SINGH NEGI
Email : manendranegi@iisc.ac.in
Designation : Phd Student
Category : Molecular and Cellular Biology

Raghavaram Peesapati
Email : raghavaramp@iisc.ac.in
Designation : Phd Student
Category : Plant and Developmental Biology
Ritabrata Basak
Email : ritabratab@iisc.ac.in
Designation : Phd Student
Category : Plant and Developmental Biology

Saurabh Bhatt
Email : saurabhbhatt@iisc.ac.in
Designation : Phd Student
Category : Plant and Developmental Biology

Shambhavi
Email : shambhavi2@iisc.ac.in
Designation : Phd Student
Category : Plant and Developmental Biology

SUSHMITA DUTTA
Email : sushmitad@iisc.ac.in
Designation : Phd Student
Category : Plant and Developmental Biology
Publications
2023
Zamzam M, Singh S, Peesapati R, Prakash S, Basak, R, Khanday I, Simonini S, Grossniklaus U, Vijayraghavan U
Unequal genetic redundancy among the rice transcription factors OsMADS2 and OsMADS4 reveals distinct roles in floret organ development, Biorxiv, 8(5)
Krishan VP, Negi MS, Peesapati R, Vijayraghavan U
Cryptococcus neoformans Slu7 ensures nuclear positioning during mitotic progression through RNA splicing, press PLoS Genetics, 11(23)
2022
Prakash S, Rai R, Mohamed ZamZam, Ahmad O, Peesapati R, Vijayraghavan U
OsbZIP47 an integrator for meristem regulators during rice plant growth and development, Frontiers in Plant Biology, 13(0)
2019
Vijayakumari D, Sharma AK, Bawa PS, Kumar R, Srinivasan S, Vijayraghavan U.
Early splicing functions of fission yeast Prp16 and its unexpected requirement for gene Silencing is governed by intronic features, RNA Biology, 16(6)
2016
Khanday I, Das S, Chongloi GL, Bansal M, Grossniklaus U, Vijayraghavan U
Genome-Wide Targets Regulated by the OsMADS1 Transcription Factor Reveals Its DNA Recognition Properties., Plant Physiol, 172(1)
2015
Deshpande GM, Ramakrishna K, Chongloi GL, Vijayraghavan U
Functions for rice RFL in vegetative axillary meristem specification and outgrowth, Journal of Experimental Botany, 66(9)
2014
Conrad L, Khanday I, Cameron J, Emmanuel G, Gynheung A, Vijayraghavan U, and Venkatesan S
The Polycomb Group Gene EMF2B is essential for maintenance of floral meristem determinacy in rice, The Plant Journal, 80(5)
2008
Rao N, Prasad K, Ravikumar P, Vijayraghavan U
A regulatory role for RFL, the rice LFY homolog, in determining flowering time and panicle architecture., PNAS, U.S.A, 105(9)
Alumni

VISHNU PRIYA KRISHNAN
vishnuk@iisc.ac.in

Dr. Vasudevan Seshadri
seshadriv@nccs.res.in

Dr. Kalika Prasad
kalika.prasad@iiserpune.ac.in

Dr. Ashis Nandi
ashis_nandi@mail.jnu.ac.in